
Division Head and Professor of Chemical Biology & Medicinal Chemistry
Romeo T. Bachand, Jr. Regents Professor in Pharmacy
Executive Committee Chair
Fellow of Alumni Centennial Endowed Fellowship in Pharmacy (P1 Teaching Award)

Our research focuses on mechanistic and structural studies of enzymes involved in the bacterial catabolism of aromatic and halogenated compounds. The specific enzymes examined under study are found in the tautomerase superfamily and the fumarylacetoacetate hydrolase (FAH) superfamily. In addition to advancing our general understanding of enzyme mechanisms, these studies provide insight into how enzymes evolve and how new activities emerge. It has become incumbent upon us to understand this process due to the ever-increasing prevalence of antibiotic-resistant bacteria and other drug-resistant organisms such as M. tuberculosis. Although there are several strategies for overcoming resistance (synthesis of new compounds, identification of new targets, inhibition of enzymes conferring resistance), a better understanding of how these new activities evolve in the first place might suggest some alternative and more effective approaches.
These studies have led us in a number of different directions. Most recently, for example, we have identified a tautomerase superfamily member (designated TomN) in a biosynthetic pathway for one ring of the anti-tumor, tomaymycin. Tomaymycin is one of the pyrrolo[1,4]benzodiazepine natural products that includes anthramycin and sibiromycin. The C ring of tomaymycin is generated in five enzyme-catalyzed steps from L-tyrosine. The genes encoding these enzymes have recently been cloned and their functions tentatively assigned, but there is limited biochemical evidence supporting the assignments of the last three steps. We are currently characterizing the enzymatic and non-enzymatic chemistry in the pathway. The ultimate goal is to use these enzymes to generate new, more potent tomaymycin analogues that are synthetically inaccessible.
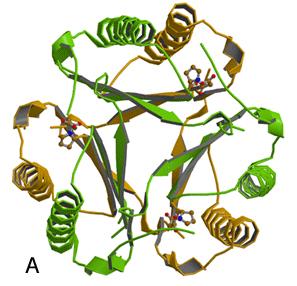
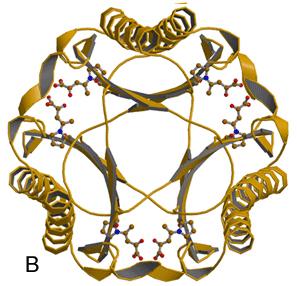
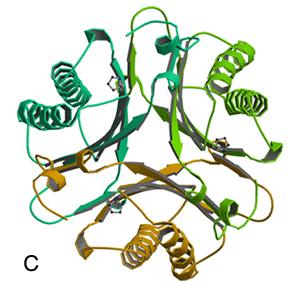
Burks, E.A., Yan, W., Johnson, Jr., W.H., Li, W., Schroeder, G.K., Min, C., Gerratana, B., Zhang, Y., and Whitman, C.P. (2011) Kinetic, crystallographic, and mechanistic characterization of TomN: elucidation of a function for a 4-oxalocrotonate tautomerase homologue in the tomaymycin biosynthetic pathway, Biochemistry 35, 7600-7611.
Schroeder, G.K., Johnson, Jr., W.H., Huddleston, J.P., Serrano, H., Johnson, K.A., and Whitman, C.P. (2012) Reaction of cis-3-chloroacrylic acid dehalogenase with an allene substrate, 2,3-butadienoate: hydration via an enamine, J. Am. Chem. Soc. 134, 293-304.
Huddleston, J.P., Schroeder, G.K., Johnson, K.A., and Whitman, C.P. (2012) A pre-steady-state kinetic analysis of the αY60W mutant of trans-3-chloroacrylic acid dehalogenase: implications for the mechanism of the wild type enzyme, Biochemistry 51, 9420-9435.
Whitman, C.P. (2013) Functional (Mis)Assignment in the Tomaymycin Biosynthetic Pathway In Experimental Standard Conditions of Enzyme Characterizations – Protein Structure Meets Enzyme Kinetics, Proceedings of the 5th International Beilstein Symposium, Rudesheim, Germany, Sept 12-16, 2011; M.G. Hicks, C. Kettner, Eds.; Beilstein-Institute, Frankfurt, Germany, 2013; 111-127.
Schroeder, G.K., Huddleston, J.P., Johnson, Jr., W.H., and Whitman, C.P. (2013) A mutational analysis of the active site loop residues in cis-3-chloroacrylic acid dehalogenase, Biochemistry 52, 4204-4216.
Guo, Y., Serrano, H., Poelarends, G.J., Johnson, Jr., W.H., Hackert, M.L., and Whitman, C.P. (2013) Kinetic, mutational, and structural analysis of malonate semialdehyde decarboxylase from Coryneform bacterium strain FG41: mechanistic implications for the decarboxylase and hydratase activities, Biochemistry 52, 4830-4831.
Terrell, C.R., Burks, E.A., Whitman, C.P., and Hoffman, D.W. (2013) Structural and kinetic characterization of two 4-oxalocrotonate tautomerases in Methylibium petroleiphilum strain PM1, Arch. Biochem. Biophys. 537, 113-124.
Poelarends, G.J., Serrano, H., Huddleston, J.P., Johnson, Jr., W.H., and Whitman, C.P. (2013) A mutational analysis of active site residues in trans-3-chloroacrylic acid dehalogenase, FEBS Lett. 587, 2842-2850.
Huddleston, J.P., Burks, E.A., and Whitman, C.P. (2014) Identification and characterization of new family members in the tautomerase superfamily, Arch. Biochem. Biophys. 564, 189-196.
Huddleston, J.P., Johnson, Jr., W.H., Schroeder, G.K., and Whitman, C.P. (2015) The accidental assignment of function in the tautomerase superfamily, Perspect. Sci. 4, 38-45.
Huddleston, J.P., Johnson, Jr., W.H., Schroeder, G.K., and Whitman, C.P. (2015) Reactions of Cg10062, a cis-3-chloroacrylic acid dehalogenase homologue, with acetylene and allene substrates: evidence for a hydration-dependent decarboxylation, Biochemistry 54, (in press).
Email address:
Phone:
Campus location:
US Mail Address:
The University of Texas at Austin
BME 6.202A
1 University Station, C0850
Austin, TX 78712-0128
FEDEX Address:
107 W. Dean Keeton St.
BME 6.202A
The University of Texas at Austin
Austin, TX 78712

